SEL : Différence entre versions
De MicMac
(→Graphic Interface) |
(→Help) |
||
| (16 révisions intermédiaires par 4 utilisateurs non affichées) | |||
| Ligne 1 : | Ligne 1 : | ||
[[Image:picto-liste.png|25px]] [[Command|List of commands]] | [[Image:picto-liste.png|25px]] [[Command|List of commands]] | ||
==Description== | ==Description== | ||
| − | SEL is an old and ugly tool, but it can help. To visualize tie points computed with Tapioca. | + | SEL is an old and ugly tool (only available on Ubuntu), but it can help. To visualize tie points computed with Tapioca. |
<br> | <br> | ||
===Syntax=== | ===Syntax=== | ||
The global syntax for SEL is | The global syntax for SEL is | ||
| − | <pre>mm3d SEL | + | <pre>mm3d SEL ImageDirectory Image1 Image2 </pre> |
| + | Note : Even if the KH argument is not mandatory, you have to fill it ! By default it is KH=NB. | ||
| + | Note : Use <code>./</code> for <i>ImageDirectory</i> if you want to use the current directory. | ||
===Help=== | ===Help=== | ||
| Ligne 20 : | Ligne 22 : | ||
* [Name=RL1] bool :: {Estimate Homography using L1 mode} | * [Name=RL1] bool :: {Estimate Homography using L1 mode} | ||
* [Name=F] string | * [Name=F] string | ||
| − | * [Name=KH] string :: {In P PB PBR M S NB NT MMD} | + | * [Name=KH] string :: {In P PB PBR M S NB NT MMD, NB stand for binary Pastis files (default)} |
* [Name=KCpl] string | * [Name=KCpl] string | ||
| − | * [Name=SzW] Pt2di | + | * [Name=SzW] Pt2di :: {Size of window} |
* [Name=ModeEpip] bool :: {If mode epip, the y displacement are forced to 0} | * [Name=ModeEpip] bool :: {If mode epip, the y displacement are forced to 0} | ||
* [Name=SH] string :: {Homologue extenion for NB/NT mode} | * [Name=SH] string :: {Homologue extenion for NB/NT mode} | ||
===Graphic Interface=== | ===Graphic Interface=== | ||
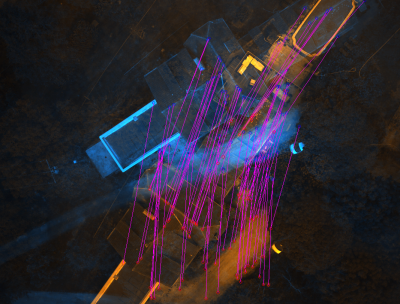
| − | SEL display a GUI with the 2 image superposed. The first one is colored in blue and the second in orange. If you click on the mouse wheel, you will see the tie points between the two images. If you click right, you will display a contextual menu : | + | SEL display a GUI with the 2 image superposed. The first one is colored in blue and the second in orange. If you click on the mouse wheel, you will see the tie points between the two images. If you click right, you will display a contextual menu :<br> |
| + | [[Image:gui_SEL.png|x300px]] | ||
*Recal | *Recal | ||
*"Polygone" : define a polygone for tie points. | *"Polygone" : define a polygone for tie points. | ||
*i1&i2 : display image1 and image2 (default). | *i1&i2 : display image1 and image2 (default). | ||
| − | *i1 : display image1. | + | *i1 : display image1 with tie points. |
| − | *i2 : display image2. | + | *i2 : display image2 with tie points. |
*"skull" : | *"skull" : | ||
*PCor | *PCor | ||
| − | *Flip | + | *Flip : switch between image1 and image2 |
| − | *Profil | + | *Profil : display image1 and image2 profil |
*Liaison | *Liaison | ||
*GR | *GR | ||
Version actuelle en date du 3 octobre 2018 à 15:24
Description
SEL is an old and ugly tool (only available on Ubuntu), but it can help. To visualize tie points computed with Tapioca.
Syntax
The global syntax for SEL is
mm3d SEL ImageDirectory Image1 Image2
Note : Even if the KH argument is not mandatory, you have to fill it ! By default it is KH=NB.
Note : Use ./ for ImageDirectory if you want to use the current directory.
Help
You can access to the help by typing :
mm3d SEL -help
Mandatory unnamed args :
- string :: {Directory}
- string :: {First image name}
- string :: {Second image name}
Named args :
- [Name=R] INT
- [Name=RL1] bool :: {Estimate Homography using L1 mode}
- [Name=F] string
- [Name=KH] string :: {In P PB PBR M S NB NT MMD, NB stand for binary Pastis files (default)}
- [Name=KCpl] string
- [Name=SzW] Pt2di :: {Size of window}
- [Name=ModeEpip] bool :: {If mode epip, the y displacement are forced to 0}
- [Name=SH] string :: {Homologue extenion for NB/NT mode}
Graphic Interface
SEL display a GUI with the 2 image superposed. The first one is colored in blue and the second in orange. If you click on the mouse wheel, you will see the tie points between the two images. If you click right, you will display a contextual menu :
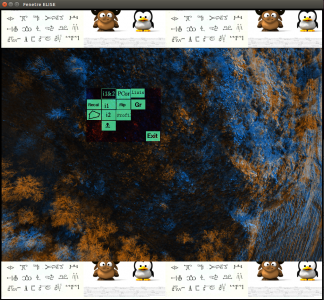
- Recal
- "Polygone" : define a polygone for tie points.
- i1&i2 : display image1 and image2 (default).
- i1 : display image1 with tie points.
- i2 : display image2 with tie points.
- "skull" :
- PCor
- Flip : switch between image1 and image2
- Profil : display image1 and image2 profil
- Liaison
- GR
- Exit : Quit.
Example
For example with the Viabon dataset :
mm3d SEL ./ image_002_00090.tif image_002_00089.tif KH=NB